Intra- and inter-cellular signaling pathways often involve chemical fluxes that are too small to detect using conventional assays and instrumentation. The Strano laboratory designs and synthesizes fluorescent nanosensors capable of listening to these signals, even at the single molecule level. Our work focuses on the synthesis and mathematical analysis of these analytical platforms to solve biological problems.
Project 1: Single-molecule Detection of Reactive Oxygen Species
Intra- and inter-cellular signaling pathways often involve chemical fluxes that are too small to detect using conventional assays and instrumentation. The Strano laboratory designs and synthesizes fluorescent nanosensors capable of listening to these signals, even at the single molecule level. Our work focuses on the synthesis and mathematical analysis of these analytical platforms to solve biological problems.
Recent advances in nanotechnology have produced the first sensor transducers capable of resolving the adsorption and desorption of single molecules. Examples include near infrared fluorescent single walled carbon nanotubes that report single molecule binding via stochastic quenching. A central question for the theory of such sensors is how to analyze stochastic adsorption events and extract the local concentration or flux of the analyte near the sensor. In this work, we compare algorithms of varying complexity for accomplishing this by first constructing a kinetic monte carlo model of molecular binding and unbinding to the sensor substrate and simulating the dynamics over wide ranges of forward and reverse rate constants. Methods involving single-site probability calculations, first and second moment analyses, and birth-and-death population modeling are compared for their accuracy in reconstructing model parameters in the presence and absence of noise over a large dynamic range. Overall, birth-and-death population modeling was the most robust in recovering the forward rate constants, with the first and second order moment analysis very efficient when the forward rate is large (> 10-3 s-1). The precision decreases with increasing noise, which we show masks the existence of underlying states. Precision is also diminished with very large forward rate constants, since the sensor surface quickly and persistently saturates.
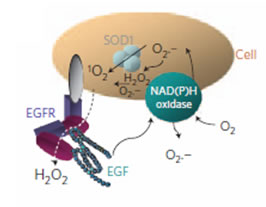
Reference: 1) Jin, H. et al. Detection of single-molecule H2O2 signalling from epidermal growth factor receptor using fluorescent single-walled carbon nanotubes. Nat Nano 5, 302-309. 2) Boghossian, A. A., Zhang, J., et al. The chemical dynamics of nanosensors capable of single-molecule detection J.Chem.Phys 135, 084124 (2011).
Project 2: Molecular Recognition Using Nanotube-Adsorbed Polymer Complexes
Molecular recognition is central to the design of therapeutics, chemical catalysis and sensor platforms, with the most common mechanisms involving biological structures such as antibodies and aptamers. The key to this molecular recognition is a folded and constrained heteropolymer pinned, via intra-molecular forces, into a unique three-dimensional orientation that creates a binding pocket or interface to recognize a specific molecule. An alternate approach to constraining a polymer in three-dimensional space involves adsorbing it onto a cylindrical nanotube surface. The Strano lab at MIT has pioneered fundamentally new types of the molecular recognition motifs based on these structured, nanotube-associated complexes. To date, we have demonstrated a growing list of examples in which natural or synthetic polymers create unique and highly selective molecular recognition sites once adsorbed onto a single-walled carbon nanotube (SWCNT) surface. The phenomenon is shown to be generic, with new recognition complexes demonstrated for nitroaromatics, riboflavin, l-thyroxine, and estradiol. We have also pioneered the first thermodynamic model to predicted adsorption and recognition into the corona of a nanoparticle adsorbed phase using a 2D thermodynamic model of surface interactions. The complexes can be used as new types of sensors based on modulation of SWCNT photoemission, as demonstrated using a complex for real time spatio-temporal detection of riboflavin in murine macrophages.
Project 3: Label-free Protein and Antibody Microarrays
Excessive sample volumes continue to be a major limitation in the analysis of protein-protein interactions, motivating the search for label-free detection methods of greater sensitivity. Herein, we report the first chemical approach for selective protein recognition using fluorescent SWNT enabling label-free microarrays capable of single protein detection. A hexahistidine-tagged capture protein is coupled to a Ni2+ chelated by N,N-bis(carboxymethyl)-L-lysine grafted to chitosan surrounding the SWNT. The Ni2+ acts as a proximity quencher with the Ni2+/SWNT distance altered upon docking of the analyte protein. This ability to discern single protein binding events decreases the apparent detection limit from 100 nM, for the ensemble average, to 10 fM for an observation time of 600 sec. This first use of cell-free synthesis to functionalize a nanosensor extends this method to an virtually infinite number of receptor proteins. To demonstrate this, the arrays are used to analyze a network of 1156 protein-protein interactions in the staurosporine-induced apoptosis of SH-SY5Y cells, confirming literature predictions.
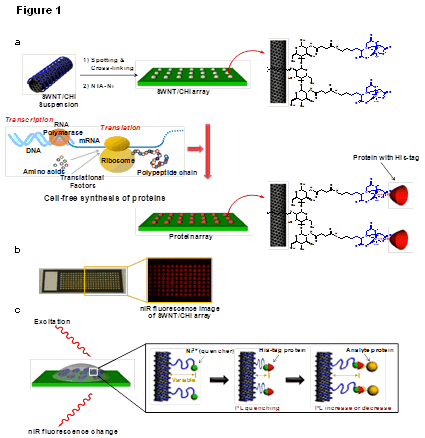
Figure: chematic of a label-free protein array based upon fluorescent single-walled carbon nanotubes. (a) Array fabrication using SWNT/CHI and in situ generation of individually addressed capture proteins using cell-free protein synthesis for label-free optical detection of protein interactions. A SWNT/CHI suspension is spotted on glass and functionalized with Ni-NTA to bind His-tag-containing capture proteins. Cell-free extract and PCR-amplified DNA coding for each protein were added to each spot for protein expression and in situ immobilization. (b) Optical and nIR fluorescence image of the SWNT/CHI array. (c) Signal transduction mechanism for label-free detection of protein-protein interactions: a nIR fluorescence change from the SWNT occurs when the distance between the Ni2+ quencher and SWNT is altered upon analyte protein binding.
Reference: Ahn, J.-H. et al. Label-Free, Single Protein Detection on a Near-Infrared Fluorescent Single-Walled Carbon Nanotube/Protein Microarray Fabricated by Cell-Free Synthesis. Nano Letters 11, 2743-2752, doi:10.1021/nl201033d (2011).