The Strano research group has introduced new synthetic techniques, such as Corona Phase Molecular Recognition, and chemical engineering physiological analysis of the human body, to solve problems in oncology, diabetes and medical imaging. This research leverages the ability of nanotechnology-enabled sensors to detect even single molecules with precision. Computationally, the ability to mathematically describe, analyze and simulate the human body has advanced with unprecedented accuracy, facilitiated in part by the Strano group research.
Research Overview
Research projects in the Strano group focus on developing advanced biomedical technologies through nanosensor platforms, computational modeling, and chemical imaging. Nanosensor Chemical Cytometry (NCC) is a technique invented by the Strano laboratory that enables rapid, non-destructive measurement of biochemical signals from living cells at the single-cell level, capturing cellular heterogeneity for applications like optimizing cellular therapeutics production. The Strano laboratory has also pioneered a next generation of computational, physiological models that describe the human body. The “Integrated Mathematical Model Mapping Performances in Animal and Clinical Trials” (IMPACT) uses a computational model of the human and animal glucoregulatory system to benchmark the efficacy of therapeutic drugs, and enables the in-silico testing of new designs, particularly for diabetes management.
The Strano laboratory is pioneering the concept of chemical imaging – spatially mapping the chemical efflux in complex tissues to gain new insight into the diseased and healthy states of soft tissues within the human body. Examples include the real time, spatial mapping of cancer biomarkers in-vivo using novel instrumentation, and leveraging the exquisite sensitivity of nanosensor technology. The goals include 3D chemical imaging for diagnosis and surgical planning. This work is in collaboration with the Koch Institute and Brigham and Women’s Hospital. Additionally, biomedical modeling and computation integrates advanced mathematical modeling to simulate physiological processes, supporting drug development and disease management for diabetes and cancer using tools like MATLAB and COMSOL. These projects collectively advance diagnostic, therapeutic, and monitoring technologies in healthcare.
Nanosensor Chemical Cytometry (NCC)
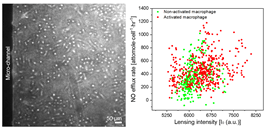
This project develops a detection platform to the study intracellular and pericellular biochemical signals from living cells and microbes in a rapid, non-destructive and label-free manner. We have recently developed Nanosensor Chemical Cytometry (NCC) that is able to measure single cell biochemical signals, allowing for the study of cellular heterogeneity within a population. This powerful approach allows for the measurement of distribution cellular states that would be lost in conventional methods. One potential application would be to monitor the quality of cellular therapeutics on a single-cell level in the process of production, allowing for dynamic optimization in the manufacturing environment. This works involves expertise in sensor engineering, microfluidics, platform development, cellular studies and computational analysis.
Reference
Mathematical Modeling of Physiological Glucose Regulation (IMPACT)
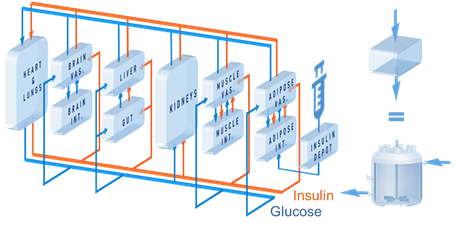
Diabetes is a disorder that alters the body’s natural glucoregulatory operation. It is known to involve complex independence of key signaling molecules such as glucose, insulin, and glucagon. This project leverages a novel computational modeling framework developed by the Strano laboratory at MIT that performs in silico simulations of the glucoregulatory system in humans and animal models such as mice, rat and minipig. The ability to make comparisons between animal and human models hold significant predictive power for drug design, such as insulin or glucagon that modulate their effectiveness as a function of glucose concentration. Recent advancements in the research area involve implementing both insulin and glucagon dynamics concurrently. We envision that the resulting algorithm will support both therapeutic discovery, clinical decision making, and personalized medicine.
Reference
Chemical Imaging
This project aims to develop a sensor platform for detecting cancer biomarkers in cell and tissue specimens. A synthetic polymer library will be used to functionalize nanoparticles, enabling selective binding of target molecules and subsequent changes in nanoparticle fluorescence. Promising sensors, characterized by their selectivity and sensitivity, will be integrated into a medical platform to map the molecular environment, providing spatial 3D chemical imaging for applications in disease diagnosis and surgical planning. This work involves both internal and external collaborations with research scientists at the Koch Institute at MIT and physicians from Brigham and Women’s Hospital at Harvard Medical School.
Reference
Biomedical Modeling and Computation
The ability to accurately predict key physiological functions is crucial for advancing the development of devices and therapeutics aimed at improving patient outcomes. Collaborating closely with experimentalists within and beyond the Strano research group, this project integrates principles of transport phenomena, numerical methods, and advanced mathematics to construct models that elucidate physiological processes of interest. Our Biomedical Modeling and Computation initiatives are used to simulate glucoregulatory systems in humans and animals using IMPACT—a pharmacokinetic model developed by SRG members. This modeling effort supports drug development and disease management for diabetes. We have also focused on analyzing biomarker transport associated with carcinogenesis in the context of cancer detection and treatment. These tasks involve the frequent use of coding languages and modeling software, such as MATLAB and COMSOL.
In Vivo Implantable Sensors
Single-walled carbon nanotubes are particularly attractive for biomedical applications, because they exhibit a fluorescent signal in a spectral region where there is minimal interference from biological media. Although single-walled carbon nanotubes have been usedas highly sensitive detectors for various compounds, their use as in vivo biomarkers requires the simultaneous optimization of various parameters, including biocompatibility, molecular recognition, high fluorescence quantum efficiency and signal transduction.
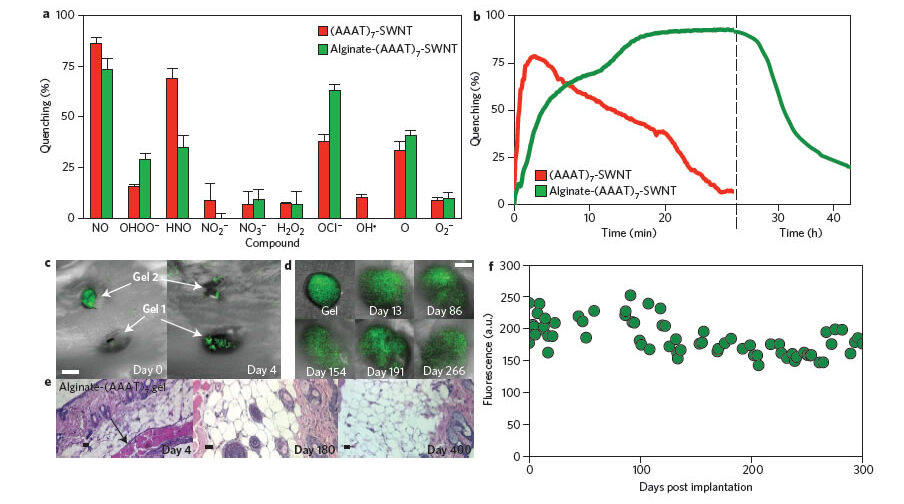
Here we show that a polyethylene glycol ligated copolymer stabilizes nearinfrared-fluorescent single-walled carbon nanotubes sensors in solution, enabling intravenous injection into mice and the selective detection of local nitric oxide concentration with a detection limit of 1 mM. The half-life for liver retention is 4 h, with sensors clearing the lungs within 2 h after injection, thus avoiding a dominant route of in vivo nanotoxicology. After localization within the liver, it is possible to follow the transient inflammation using nitric oxide as a marker and signalling molecule.
To this end, we also report a spatial-spectral imaging algorithm to deconvolute fluorescence intensity and spatial information from measurements. Finally, we demonstrate that alginate-encapsulated single-walled carbon nanotubes can function as implantable inflammation sensors for nitric oxide detection, with no intrinsic immune reactivity or other adverse response for more than 400 days.